Research
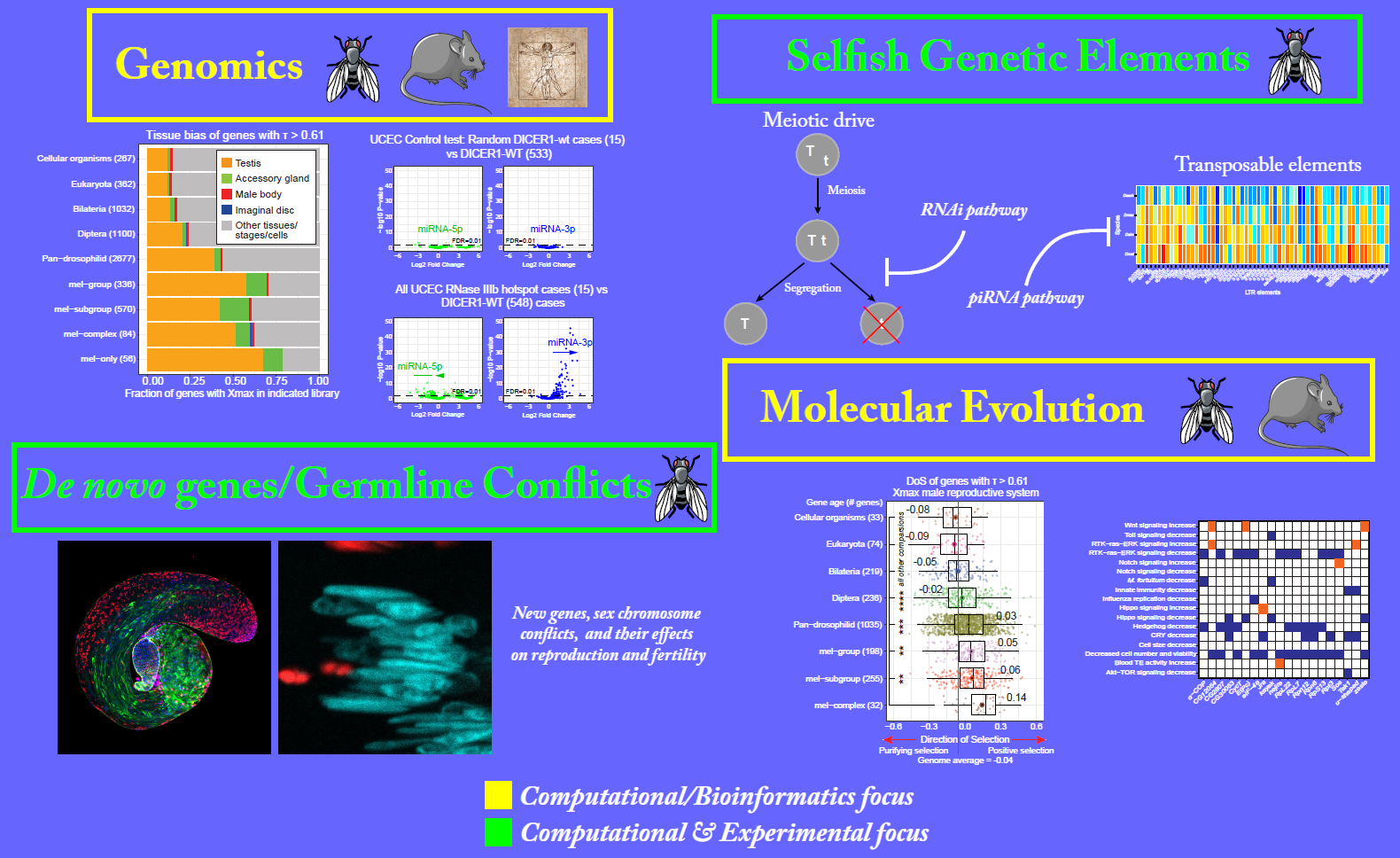
Welcome to the Evolutionary and Developmental Genomics (EDGe) lab at UTSA. Our group is nested within the Developmental and Regenerative Sciences program in the Department of Neuroscience, Developmental and Regenerative Biology. Broadly, our research focuses on understanding the evolution and function(s) of a class of ‘selfish’ genes called meiotic drive genes, which violate Mendel’s law of segregation during gamete formation (sperm and egg cells). We study how their selfish activities can adversely affect germline development, reproduction, and fertility. We use Drosophila as a model study meiotic drive, and our research utilizes a combination of evolutionary genomics, molecular biology, and bioinformatics approaches to study their molecular mechanisms of action.
Besides, our laboratory is also interested in understanding how genetic conflicts mediated by selfish genes (such as meiotic drive genes and transposable elements) shape the evolution of genes essential for germline development and fertility. On a connected theme, using extensive integrative genomic resources available from model organisms such as Drosophila, mice, and primates, our research focuses on discovering novel genes (conserved and lineage-specific) essential for male fertility.


Jeffrey Vedanayagam PhD
Principal Investigator
jeffrey.vedanayagam@utsa.edu
I joined UTSA’s Developmental and Regenerative Sciences program within the Department of Neuroscience, Developmental and Regenerative Biology as an Assistant Professor in Fall 2023. Prior to that, I was an NIH Pathway to Independence postdoctoral fellow in the Developmental Biology Program at the Memorial Sloan Kettering Cancer Center in New York and obtained my Ph.D. in Evolutionary/Population genomics from the University of Rochester, NY. I began to use Drosophila as a model to study germline conflicts both during my graduate and postdoctoral work. At UTSA, our work on selfish genes and genetic conflicts continues with the Drosophila model. In addition, we use bioinformatics approaches to examine genome sequence data from vertebrate model organisms such as mice to understand how selfish genes shape genome evolution.

Jodh Singh Pannu PhD
Associate Research Scientist
jodh.pannu@utsa.edu
..

Milagros "Mili" Alonso
Lab Manager
milagros.alonso@utsa.edu
My name is Milagros Alonso, Mili for short, and I was born in Mar del Plata, Argentina. I obtained my Bachelors degree at UTSA in Environmental Science with a concentration in Wildlife Management and conservation. Since my graduation from UTSA, I’ve had the pleasure of working with and learning from some of the most intelligent and passionate Keepers in the zoological field at the San Antonio Zoo, and a non-profit animal education facility called Zoomagination. Now, by joining UTSA’s own Neuroscience, Developmental and Regenerative Biology Department as a Research Assistant Scientist, I’m excited to return to my roots to push my learning and understanding of the Sciences that govern our lives every day.

Kendall Green
PhD Candidate
kendall.green@utsa.edu
My name is Kendall Green and I was born and raised in Dallas, Texas. I graduated from Texas A&M University in 2019 with a Bachelor’s of Science in Biomedical Sciences. I received my Master of Science in Biology in 2021 from University of Houston. After I graduated, I worked as a Research Assistant at Baylor College of Medicine’s Human Genome Sequencing Center, which further ignited my interests in molecular biology and genomics and inspired me to pursue a PhD. When I am not in lab doing experiments, my hobbies include knitting and crocheting and caring for two aquariums--a 10 gallon with tropical fish and an albino catfish and a 29-gallon housing a beautiful chunky goldfish. I also enjoy working out, reading, cooking, and listening to music. I absolutely love spending time with my family. I have one older sister and one younger sister, and we love making our parents laugh. We are all very close and they have always supported my passion for science.

Joel Osegueda-Delgado
PhD Candidate
joel.oseguedadelgado@utsa.edu
My name is Joel Osegueda Delgado and I was born in Phoenix, Arizona, raised in Houston, Texas. I graduated from the University of Houston-Downtown with a B.S. in Biotechnology with dual concentration in microbiology and chemistry. In my time as an undergraduate, I worked doing guided research understanding sleep and alcohol tolerance in fruit flies. That opportunity took me to a lab that worked on the blood brain barrier in flies while working independently as a mentor for highschool students. These experiences bolstered my fascination of the mechanisms that gives us function and thus took me to a PhD in the Developmental and Regenerative Sciences PhD program. Outside of lab, my time is full of scouring the internet for a new recipe to try, new music to fill the air, or a new place to visit for lunch/sightseeing. Additionally, one of my favorite things to do is create new 3D designs to make things more accessible for others and make life easier.

Cassandra Spondike
Research Science Associate
Works with Joel
I am a senior majoring in Microbiology and Immunology here at UTSA. During my time here, I’ve realized my interest in research and I hope to pursue graduate school and a career in infectious disease after completing my Bachelors. In my free time, I love hiking, swimming, reading, or playing games with my friends and family. I’m so grateful for the opportunity to get hands on research experience in this lab

Daisy Kolo
Research Science Associate
Works with Mili
..

Xander Zamora
Undergrad volunteer
Works with Joel
..

Zeno
Lab Mascot
I am Zeno and I like paradoxes. I provide administrative (and emotional) support to the EDGe lab. I was rescued in Brooklyn, NY, and made my way to Texas when the lab was founded in Fall 2023. I have mastered the art of sighing and each sigh is loaded with emotion. Especially, when you show your rejected papers, grants, summary statements, etc. You can call me the Shakespeare of sighs. Make sure you don't drop anything on the floor, I am on it faster than any high-tech vacuum. Also remember, I'm eco-friendly and powered entirely by treats and belly rubs. I am here to cheer you all up!

Talha Mazhar
Undergrad volunteer
I was born in Chicago, however I moved to San Antonio after my first birthday. I have two sisters, one younger and one older, and a total of 5 cats! I love all of them equally, and honestly would not be able to live without them, they mean the world to me. Fortunately, I am able to pursue higher education for something that I love, which is medicine. At my time at college, I was fortunate to be able to meet some amazing new people and form plenty of new friendships, which allowed me to try out different things that I normally wouldn’t such as learning to play a piano or competitively play chess. Throughout my time in college, I also want to be able to contribute my time to advance research in any possible way in order to continue to improve people’s health. Some of my all-time favorite activities/hobbies include traveling, listening to new music, boxing, reading, hiking, and playing games. My ultimate goal is to go to medical school to become an anesthesiologist, and give back to my parents and the community.

Sofia Sanchez
Undergrad volunteer
I am currently in my senior year, with interest in stem cell, regenerative science, and virology research. Following the completion of my undergraduate studies, I plan to pursue a career in the Air Force as a biomedical research scientist for a couple years then go back to school to achieve a PhD. Outside of my academic and professional pursuits, I enjoy traveling, hiking, and thrifting

Malaika Koreshi
Undergrad volunteer
My name is Malaika Koreshi and I am an undergraduate student at UTSA. As of this summer 2024, I have completed two semesters with UTSA. I am a psychology major and am going to pursue a biology minor with the intention of going pre-med. I have lived in San Antonio for almost 12 years. I love to read and draw and I absolutely love watching nature documentaries. Fun fact about me, my family and I have two backyard chickens named Coco and Alice!
The Escape Room
Our first lab outing and doing the easiest room was still a challenge! Still escaped!


Peer-reviewed publications
14. Signor. S, Vedanayagam. J, Kim. B. Y, Wierzbicki. F, Kofler. R, Lai. E.C. Rapid evolutionary diversification of the flamenco locus across simulans clade Drosophila species PLoS Genetics 2023; 19 (8): e1010914. PDF
13. Vedanayagam. J, Lin. C, Papareddy. R, Nodine. M, Flynt A.S, Wen. J, Lai. E.C. Regulatory logic of endogenous RNAi in silencing de novo genomic conflicts PLoS Genetics 2023; 19 (6): e1010787. PDF
12. Vedanayagam. J*, Herbette. M*, Mudgett. H, Lin. C, Gunasinghe. H, McDonough-Goldstein. C, Dorus. S, Loppin. B, Meiklejohn. C, Dubruille. R, Lai. E.C. Essential and recurrent roles for hairpin RNAs in silencing de novo sex chromosome conflict in Drosophila simulans PLoS Biology 2023; 21 (6): e3002136. *equal contribution PDF
11. Shang. R, Kretov. D.R, Adamson. I.S, Treiber. T, Treiber. N, Vedanayagam. J, Chuang. J.H, Meister. G, Cifuentes. D, Lai. E.C. Regulated dicing of pre-mir-144 via reshaping of its terminal loop . Nucleic Acids Res. 2022; 50 (13): 7637-7654. PDF
10. Vedanayagam. J, Lin. C, Lai. E.C. Rapid evolutionary dynamics of an expanding family of meiotic drive factors and their hpRNA suppressors . Nature Ecol. & Evol. 2021; 5: 1613–1623. PDF
9. Chakraborty. M, Chang. C-H, Khost. D.E, Vedanayagam. J, Adrion. J.R, Liao. Y, Montooth. K, Meiklejohn. C.D, Larracuente. A.M, Emerson. J.J. Evolution of genome structure in the Drosophila simulans species complex . Genome Res. 2021; 31:1-17. PDF
8. Vedanayagam. J, Chatila. W, Aksoy. B.M, Majumdar. S, Skanderup. A.J, Lee. W, Demir. E, Ciriello. G, Schultz. N, Sander. C, Lai. E.C. Cancer-associated mutations in DICER1 RNase IIIa and IIIb domains exert similar effects on miRNA biogenesis. Nature Commun., 2019; 10:3682.PDF
7. Meiklejohn. C.D, Landeen. E.L, Gordon. K.E, Rzatkiewicz. T, Kingan. S.B, Geneva. A.J, Vedanayagam. J, Muirhead.C.A, Garrigan. D, Stern. D.L, Presgraves. D.C. Gene flow mediates the role of sex chromosome meiotic drive during complex speciation. eLife 2018; 7:e354. PDF
6. Lin. C, Hu. F, Dubruille. R, Vedanayagam. J, Wen. J, Smibert. P, Loppin. B, Lai. E.C. The hpRNA/RNAi pathway is essential to resolve intragenomic conflict in the Drosophila male germline. Developmental Cell 2018; 46: 316-326. PDF
5. Kondo.S*, Vedanayagam. J*, Mohammed. J, Eizadshenass. S, Kan. L, Pang. N, Aradhya. R, Siepel. A, Steinhauer. J, Lai. E.C. New genes often acquire male-specific functions but rarely become essential in Drosophila . Genes & Development 2017; 31:1841-1846. *equal contribution PDF
4. Kan.L, Grozhik A.V, Vedanayagam. J, Patil D.P, Pang. N, Lim. K, Huang. Y, Joseph. B, Lin. C, Despic. V, Guo. J, Yan. D, Kondo. S, Deng. W, Dedon. P, Jaffrey. S.R, Lai. E.C. The m6A pathway facilitates sex determination in Drosophila. Nature Commun. 2017; 8:15737. PDF
3. Vedanayagam. J, Garrigan. D. The effects of natural selection across molecular pathways in Drosophila melanogaster . BMC Evol Biol. 2015; 15:203.PDF
2. Garrigan. D, Kingan S.B, Geneva. A.J, Vedanayagam. J, Presgraves. D.C. Genome diversity and divergence in Drosophila mauritiana: multiple signatures of faster X evolution . Genome Biol Evol. 2014; 6(9):2444–2458. PDF
1. Nandineni. M.R, Vedanayagam. J. Selective enrichment of human DNA from non-human DNAs for DNA typing of decomposed skeletal remains . Forensic Sci.Int. Gene. Suppl. 2009; 2(1) 520-521. PDF
National Institute of General Medical Sciences awarded a K99/R00 funding for my work to study evolutionary and molecular bases of meiotic drive and suppression. Huge thanks to people who have supported me in this endeavor of beginner’s grantsmanship.
Ongoing support:

R00 Pathway to Independence Award (2024-2026)
Completed support:

NSF Doctoral Dissertation Improvement Grant (2012-2015)